Reducing Python String Memory Use in Apache Arrow 0.12
Published
05 Feb 2019
By
Wes McKinney (wesm)
Python users who upgrade to recently released pyarrow 0.12 may find that
their applications use significantly less memory when converting Arrow string
data to pandas format. This includes using pyarrow.parquet.read_table and
pandas.read_parquet. This article details some of what is going on under the
hood, and why Python applications dealing with large amounts of strings are
prone to memory use problems.
Why Python strings can use a lot of memory
Let’s start with some possibly surprising facts. I’m going to create an empty
bytes object and an empty str (unicode) object in Python 3.7:
In [1]: val = b''
In [2]: unicode_val = u''
The sys.getsizeof function accurately reports the number of bytes used by
built-in Python objects. You might be surprised to find that:
In [4]: import sys
In [5]: sys.getsizeof(val)
Out[5]: 33
In [6]: sys.getsizeof(unicode_val)
Out[6]: 49
Since strings in Python are nul-terminated, we can infer that a bytes object
has 32 bytes of overhead while unicode has 48 bytes. One must also account for
PyObject* pointer references to the objects, so the actual overhead is 40 and
56 bytes, respectively. With large strings and text, this overhead may not
matter much, but when you have a lot of small strings, such as those arising
from reading a CSV or Apache Parquet file, they can take up an unexpected
amount of memory. pandas represents strings in NumPy arrays of PyObject*
pointers, so the total memory used by a unique unicode string is
8 (PyObject*) + 48 (Python C struct) + string_length + 1
Suppose that we read a CSV file with
- 1 column
- 1 million rows
- Each value in the column is a string with 10 characters
On disk this file would take approximately 10MB. Read into memory, however, it
could take up over 60MB, as a 10 character string object takes up 67 bytes in a
pandas.Series.
How Apache Arrow represents strings
While a Python unicode string can have 57 bytes of overhead, a string in the Arrow columnar format has only 4 (32 bits) or 4.125 (33 bits) bytes of overhead. 32-bit integer offsets encodes the position and size of a string value in a contiguous chunk of memory:
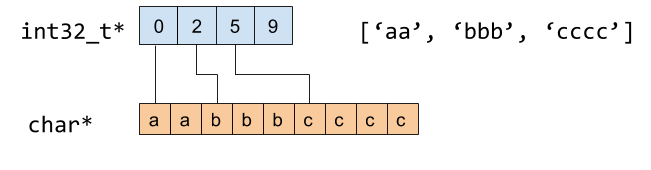
When you call table.to_pandas() or array.to_pandas() with pyarrow, we
have to convert this compact string representation back to pandas’s
Python-based strings. This can use a huge amount of memory when we have a large
number of small strings. It is a quite common occurrence when working with web
analytics data, which compresses to a compact size when stored in the Parquet
columnar file format.
Note that the Arrow string memory format has other benefits beyond memory use. It is also much more efficient for analytics due to the guarantee of data locality; all strings are next to each other in memory. In the case of pandas and Python strings, the string data can be located anywhere in the process heap. Arrow PMC member Uwe Korn did some work to extend pandas with Arrow string arrays for improved performance and memory use.
Reducing pandas memory use when converting from Arrow
For many years, the pandas.read_csv function has relied on a trick to limit
the amount of string memory allocated. Because pandas uses arrays of
PyObject* pointers to refer to objects in the Python heap, we can avoid
creating multiple strings with the same value, instead reusing existing objects
and incrementing their reference counts.
Schematically, we have the following:
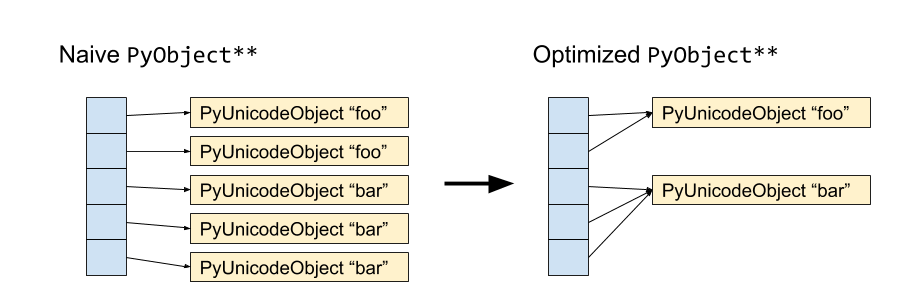
In pyarrow 0.12, we have implemented this when calling to_pandas. It
requires using a hash table to deduplicate the Arrow string data as it’s being
converted to pandas. Hashing data is not free, but counterintuitively it can be
faster in addition to being vastly more memory efficient in the common case in
analytics where we have table columns with many instances of the same string
values.
Memory and Performance Benchmarks
We can use the memory_profiler Python package to easily get process
memory usage within a running Python application.
import memory_profiler
def mem():
return memory_profiler.memory_usage()[0]
In a new application I have:
In [7]: mem()
Out[7]: 86.21875
I will generate approximate 1 gigabyte of string data represented as Python
strings with length 10. The pandas.util.testing module has a handy rands
function for generating random strings. Here is the data generation function:
from pandas.util.testing import rands
def generate_strings(length, nunique, string_length=10):
unique_values = [rands(string_length) for i in range(nunique)]
values = unique_values * (length // nunique)
return values
This generates a certain number of unique strings, then duplicates then to yield the desired number of total strings. So I’m going to create 100 million strings with only 10000 unique values:
In [8]: values = generate_strings(100000000, 10000)
In [9]: mem()
Out[9]: 852.140625
100 million PyObject* values is only 745 MB, so this increase of a little
over 770 MB is consistent with what we know so far. Now I’m going to convert
this to Arrow format:
In [11]: arr = pa.array(values)
In [12]: mem()
Out[12]: 2276.9609375
Since pyarrow exactly accounts for all of its memory allocations, we also
check that
In [13]: pa.total_allocated_bytes()
Out[13]: 1416777280
Since each string takes about 14 bytes (10 bytes plus 4 bytes of overhead), this is what we expect.
Now, converting arr back to pandas is where things get tricky. The minimum
amount of memory that pandas can use is a little under 800 MB as above as we
need 100 million PyObject* values, which are 8 bytes each.
In [14]: arr_as_pandas = arr.to_pandas()
In [15]: mem()
Out[15]: 3041.78125
Doing the math, we used 765 MB which seems right. We can disable the string
deduplication logic by passing deduplicate_objects=False to to_pandas:
In [16]: arr_as_pandas_no_dedup = arr.to_pandas(deduplicate_objects=False)
In [17]: mem()
Out[17]: 10006.95703125
Without object deduplication, we use 6965 megabytes, or an average of 73 bytes per value. This is a little bit higher than the theoretical size of 67 bytes computed above.
One of the more surprising results is that the new behavior is about twice as fast:
In [18]: %time arr_as_pandas_time = arr.to_pandas()
CPU times: user 2.94 s, sys: 213 ms, total: 3.15 s
Wall time: 3.14 s
In [19]: %time arr_as_pandas_no_dedup_time = arr.to_pandas(deduplicate_objects=False)
CPU times: user 4.19 s, sys: 2.04 s, total: 6.23 s
Wall time: 6.21 s
The reason for this is that creating so many Python objects is more expensive than hashing the 10 byte values and looking them up in a hash table.
Note that when you convert Arrow data with mostly unique values back to pandas, the memory use benefits here won’t have as much of an impact.
Takeaways
In Apache Arrow, our goal is to develop computational tools to operate natively on the cache- and SIMD-friendly efficient Arrow columnar format. In the meantime, though, we recognize that users have legacy applications using the native memory layout of pandas or other analytics tools. We will do our best to provide fast and memory-efficient interoperability with pandas and other popular libraries.